Pretty much entirely based on this paper: Electromagnetic extension of the Dory– Guest–Harris instability as a benchmark for Vlasov–Maxwell continuum kinetic simulations of magnetized plasmas
Closed Integral Form of Dispersion Relation#
Iman’s done a great job deriving a closed-form integral representation of the dispersion relation for the Dory-Guest-Harris instability. The trick to computing solutions to the dispersion relation is in getting all of the the normalization correct and computing the correct quadrature across both integrals (over $v_\perp$ and $\theta$).
The DGH dispersion relation is derived by perturbing Vlasov-Maxwell system about a spatially uniform equilibrium state \( f_s ^0 (v) \) in a uniform magnetic field \( B^0 \), leading to equilibrium cyclotron motion. We linearize the Vlasov-Maxwell system and analyze the equilibrium response.
Let \( B_0 \) be in the \( \hat{z} \) direction. We work in a cylindrical coordinate system such that
$$ \vec v = v_\perp \cos \phi \hat{x} + v_\perp \sin \phi \hat{y} + v_\parallel \hat{z} $$
Without loss of generality, we rotate the coordinate frame such that
$$ \vec k = k_\perp \hat{x} + k_\parallel \hat{z} $$
The \( v_\parallel \) component de-couples, and we are not interested, so let \(v_\parallel = 0\). Would need to be re-introduced for e.g. 2D3V or 3D3V situation, but we will only consider 1D2V here.
After some hard-fought applied mathematics, we get the electromagnetic dispersion relation as
$$ D(\omega, k_\perp) = K_{11} \left( K_{22} - \frac{(\omega_p \tau)^2 k_\perp ^2}{(\omega_c \tau)^2 \omega ^2} \right) - K_{12} K_{21} $$
where
$$ K_{11} = 1 + \chi_{11} $$
$$ K_{12} = \chi_{12} $$
$$ K_{21} = \chi_{21} $$
$$ K_{22} = 1 + \chi_{22} $$
\[\chi = - \frac{2 \pi (\omega_p \tau)^2}{\omega (\omega_c \tau)} \sum_s \frac{\omega_{p, s} ^2}{\omega_{c, s}} \sum_{n = - \infty}^{\infty} \int_0 ^{\infty} \frac{\partial F_s ^0}{\partial v_\perp} \begin{bmatrix} \frac{n^2}{\beta_s ^2} J_n ^2 (\beta_s) & i \frac{n}{\beta_s} J_n (\beta_s) J_n ^\prime (\beta_s) \\\\ - i \frac{n}{\beta_s} J_n(\beta_s) J_n ^\prime(\beta_s) & J_n ^\prime (\beta_s) J_n ^\prime (\beta_s) \end{bmatrix} \frac{v_\perp ^2}{n - \alpha _s} \, d v_\perp\]$$ \alpha _s \equiv \frac{\omega}{(\omega _c \tau) \omega _{c, s}} $$
$$ \beta_s \equiv \frac{k_\perp v_\perp}{(\omega_c \tau) \omega_{c, s}} $$
$$ F_s ^0 \equiv \frac{f_s ^0}{n_s} $$
$$ \omega_{p, s}^2 \equiv \frac{Z_s ^2 n_s }{A_s} $$
$$ \omega_{c, s} \equiv \frac{Z_s}{A_s} B^0 $$
Manipulation using Lerche-Newberger sum rule, we can get
\[\chi = -\frac{2 \pi (\omega_p \tau)^2}{\omega (\omega_c \tau)} \sum_s \frac{\omega_{p, s}^2}{\omega_{c, s}} \int_0 ^\infty \frac{\partial F_s ^0 }{\partial v_\perp} \begin{bmatrix} \frac{\alpha_s}{\beta_s ^2} (1 - Q_s) & - \frac{i}{2 \beta_s} Q_s ^\prime \\ \frac{i}{2 \beta_s} Q_s ^\prime & - \left( \frac{\pi}{\sin (\pi \alpha_s)} J_{- \alpha_s} ^\prime (\beta_s) J_{\alpha_s} ^\prime (\beta_s) + \frac{\alpha_s}{\beta_s ^2} \right) \end{bmatrix} v_\perp ^2 \, d v_\perp\]where
$$ Q_s \equiv \frac{\pi \alpha_s}{\sin (\pi \alpha_s)} J_{- \alpha_s }(\beta_s) J_{\alpha_s}(\beta_s) $$
$$ Q_s ^\prime \equiv \frac{\pi \alpha_s}{\sin (\pi \alpha_s)} \frac{\partial}{\partial \beta_s} \left[ J_{- \alpha_s}(\beta_s) J_{\alpha_s}(\beta_s) \right] $$
We can re-cast the Bessel function products in terms of integrals of real low-order Bessel functions:
$$ J_{- \alpha_s}(\beta) J_{\alpha_s}(\beta) = \frac{2}{\pi} \int_0 ^{\pi / 2} J_0 (2 \beta \cos \theta) \cos (2 \alpha_s \theta) d \theta $$
$$ \frac{\partial}{\partial \beta} [J_{-\alpha_s} (\beta) J_{\alpha_s} (\beta)] = - \frac{4}{\pi} \int_0 ^{\pi / 2} J_1 (2 \beta \cos \theta) \cos \theta \cos (2 \alpha_s \theta) d \theta $$
$$ J^\prime _{- \alpha_s} (\beta) J^\prime _{\alpha_s} (\beta_s) = \frac{1}{\pi} \left[ \int_0 ^{\pi / 2} \cos (2 \alpha_s \theta) [J_2 (2 \beta \cos \theta) - J_0(2 \beta \cos \theta) \cos (2 \theta)] d \theta - \frac{\alpha_s}{\beta^2} \sin (\pi \alpha_s)\right] $$
We are looking at a single-species picture, and we choose electrons. That means that
$$ A_s = 1 $$ $$ Z_s = -1 $$ $$ n = 1 $$
We choose (the cases in https://doi.org/10.1016/j.jcp.2014.08.014):
$$ B^0 = 1 $$ $$ v_\perp ^0 = \sqrt{2} $$ $$ f^0 (v_\perp) \equiv \frac{1}{\pi \alpha_\perp ^2 j!} \left(\frac{v_\perp ^2}{\alpha_\perp ^2} \right)^j e^{- v_\perp ^2 / \alpha_\perp ^2} $$
Things I need for integration:#
$$ (\omega_p \tau) = 1 $$ $$ (\omega_c \tau) = (\omega_p / \omega_c)^{-1} $$ $$ \omega = \text{(free parameter)} $$ $$ \omega_{p_s} \equiv \frac{Z_s ^2 n_s}{A_s} = 1 $$ $$ \omega_{c_s} \equiv \frac{Z_s}{A_s} B^0 = -1 $$
\[\partial F^0 _s / \partial v_\perp = \frac{1}{\pi \alpha_\perp ^2 j!} \frac{2 v_\perp}{\alpha_\perp ^2} e^{- v_\perp ^2 / \alpha_\perp ^2} \left( \frac{v_\perp ^2}{\alpha_\perp ^2}\right)^{j-1} \left[j - \frac{v_\perp ^2}{\alpha_\perp ^2} \right]\]$$ n = 1 $$
$$ \omega_p / \omega_c = \left(\frac{n_0 m_0}{\epsilon_0 B_0 ^2} \right)^{1/2} = \text{(free parameter)} $$
($\omega_p / \omega_c$ dictates the relative strength of $B$ to $E$)
We express $k_\perp$ in terms of a normalized wavenumber $\tilde{k}$:
$$ \tilde{k} \equiv k_\perp v_{\perp, 0} \frac{\omega_p}{\omega_c} \quad \text{(free parameter)} $$
$$ \rightarrow \beta_s = - \frac{\tilde{k} v_{\perp}}{v_{\perp, 0}} $$
Now let’s try to factor out some of the mess from $\chi$. Substituting in our single-species values for $(\omega_{p, s})$, $(\omega_{c, s})$, $(\omega_p \tau)$, and $(\omega_c \tau)$:
\[\begin{aligned} \chi & = & -\frac{2 \pi (\omega_p \tau)^2}{\omega (\omega_c \tau)} \frac{\omega_{p, s}^2}{\omega_{c, s}} \int_0 ^\infty \frac{\partial F_s ^0 }{\partial v_\perp} v_\perp ^2 \begin{bmatrix} \frac{\alpha_s}{\beta_s ^2} (1 - Q_s) & - \frac{i}{2 \beta_s} Q_s ^\prime \\ \frac{i}{2 \beta_s} Q_s ^\prime & - \left( \frac{\pi}{\sin (\pi \alpha_s)} J_{- \alpha_s} ^\prime (\beta_s) J_{\alpha_s} ^\prime (\beta_s) + \frac{\alpha_s}{\beta_s ^2} \right) \end{bmatrix} \, d v_\perp \\ & = & -\frac{2 \pi (\omega_p \tau)^2}{\omega (\omega_c \tau)} \frac{\omega_{p, s}^2}{\omega_{c, s}} \int_0 ^\infty \frac{\partial F_s ^0 }{\partial v_\perp} \frac{v_\perp ^2}{\beta_s ^2} \beta_s ^2 \begin{bmatrix} \frac{\alpha_s}{\beta_s ^2} (1 - Q_s) & - \frac{i}{2 \beta_s} Q_s ^\prime \\ \frac{i}{2 \beta_s} Q_s ^\prime & - \left( \frac{\pi}{\sin (\pi \alpha_s)} J_{- \alpha_s} ^\prime (\beta_s) J_{\alpha_s} ^\prime (\beta_s) + \frac{\alpha_s}{\beta_s ^2} \right) \end{bmatrix} \, d v_\perp \\ & = & -\frac{2 \pi (\omega_p \tau)^2}{\omega (\omega_c \tau)} \frac{\omega_{p, s}^2}{\omega_{c, s}} \frac{(\omega_c \tau)^2 \omega_{c, s}^2}{k_\perp ^2} \int_0 ^\infty \frac{\partial F_s ^0 }{\partial v_\perp} \beta_s ^2 \begin{bmatrix} \frac{\alpha_s}{\beta_s ^2} (1 - Q_s) & - \frac{i}{2 \beta_s} Q_s ^\prime \\ \frac{i}{2 \beta_s} Q_s ^\prime & - \left( \frac{\pi}{\sin (\pi \alpha_s)} J_{- \alpha_s} ^\prime (\beta_s) J_{\alpha_s} ^\prime (\beta_s) + \frac{\alpha_s}{\beta_s ^2} \right) \end{bmatrix} \, d v_\perp \\ & = & -\frac{2 \pi v_{\perp, 0}^2 (\omega_p / \omega_c)}{\tilde{k}^2 \omega} \int_0 ^\infty \frac{\partial F_s ^0 }{\partial v_\perp} \beta_s ^2 \begin{bmatrix} \frac{\alpha_s}{\beta_s ^2} (1 - Q_s) & - \frac{i}{2 \beta_s} Q_s ^\prime \\ \frac{i}{2 \beta_s} Q_s ^\prime & - \left( \frac{\pi}{\sin (\pi \alpha_s)} J_{- \alpha_s} ^\prime (\beta_s) J_{\alpha_s} ^\prime (\beta_s) + \frac{\alpha_s}{\beta_s ^2} \right) \end{bmatrix} \, d v_\perp \\ & = & -\frac{2 \pi v_{\perp, 0}^2 (\omega_p / \omega_c)}{\tilde{k}^2 \omega} \int_0 ^\infty \frac{\partial F_s ^0 }{\partial v_\perp} \begin{bmatrix} \alpha_s (1 - Q_s) & - \frac{i \beta_s}{2} Q_s ^\prime \\ \frac{i \beta_s}{2} Q_s ^\prime & - \left( \frac{\pi \beta_s ^2}{\sin (\pi \alpha_s)} J_{- \alpha_s} ^\prime (\beta_s) J_{\alpha_s} ^\prime (\beta_s) + \alpha_s \right) \end{bmatrix} \, d v_\perp \\ \end{aligned}\]Let’s re-write things a bit for simplicity:
\[R_{ij} = \begin{bmatrix} \alpha _s (1 - Q _s) & - \frac{i \beta _s}{2} Q ^\prime _s \\\\ \frac{i \beta _s}{2} Q^\prime _s & - \beta _s ^2 \left( \frac{\pi}{\sin (\pi \alpha _s)} J_{- \alpha _s} ^\prime (\beta _s) J_{\alpha _s} ^\prime (\beta _s) + \frac{\alpha _s}{\beta _s ^2} \right) \end{bmatrix}\]Using Bessel function identities,
$$ R_{22} = - \beta_s ^2 \left( \frac{\pi}{\sin (\pi \alpha_s)} J_{- \alpha_s} ^\prime (\beta_s) J_{\alpha_s} ^\prime (\beta_s) + \frac{\alpha_s}{\beta_s ^2} \right) \\ = - \beta_s ^2 \left(\frac{\pi}{\sin (\pi \alpha_s)} \left[ \frac{1}{\pi} \left( \int _0 ^{\pi / 2} \cos (2 \alpha_s \theta) \left[ J_2(2 \beta_s \cos \theta) - J_0 (2 \beta_s \cos \theta) \cos (2 \theta) \right] d \theta - \frac{\alpha_s}{\beta_s ^2} \sin (\pi \alpha_s) \right) \right] + \frac{\alpha_s}{\beta_s} \right) \\ = - \frac{\beta_s ^2}{\sin (\pi \alpha_s)} \int_0 ^{\pi / 2} \cos (2 \alpha_s \theta) \left[ J_2(2 \beta_s \cos \theta) - J_0 (2 \beta_s \cos \theta) \cos (2 \theta) \right] \, d \theta $$
Still not pretty, but we can give it a try. Let’s try using https://github.com/JuliaMath/QuadGK.jl to compute the improper integral over $v_\perp$, and use Gauss-Legendre quadrature to compute the interior $\theta$ integrals.
using Pkg
Pkg.add("Plots")
Pkg.add("PythonPlot")
Pkg.add("HDF5")
Pkg.add("Optim")
Pkg.add("LaTeXStrings")
Pkg.add(url="https://github.com/JuliaApproximation/FastGaussQuadrature.jl")
Pkg.add(url="https://github.com/JuliaMath/Bessels.jl.git")
Pkg.add(url="https://github.com/JuliaMath/QuadGK.jl")
using Bessels # besselj
using FastGaussQuadrature # compute gauss-legendre points
using HDF5 # store/read from file
using LaTeXStrings # Plot formatting
using LinearAlgebra # dot.()
using Optim # Newton-Raphson descent to find zeros
using Plots # Matplotlib metapackage
using QuadGK # Alternative integrator
# Set plotting backend to matplotlib
pythonplot()
# Set plotting backend to GR
# gr()
# Base.@kwdef is a macro that allows for keyword arguments and default values
Base.@kwdef struct DispersionCase
id::String
knorm::Float64
j::Int
width::Float64
wpwc::Float64
guesses::Vector{Vector{Float64}}
end
const case_a = DispersionCase(id="A", knorm=3.15, j=6, width=sqrt(1/3), wpwc=20.0, guesses=[[0.0,0.5]])
const case_b = DispersionCase(id="B", knorm=4.64, j=6, width=sqrt(1/3), wpwc=20.0, guesses=[[1.0, 0.3]])
const case_c = DispersionCase(id="C", knorm=2.12, j=2, width=1.0, wpwc=10.0, guesses=[[0.8, 0.12]])
const case_d = DispersionCase(id="D", knorm=2.12, j=1, width=sqrt(2), wpwc=1.0, guesses=[[1.5, 0.0]])
const cases = [case_a, case_b, case_c, case_d]
function solve_em_dispersion(;knorm, j, width, wpwc, wr, wi)
v_0 = sqrt(j) * width
function dF0_dv(v)
return 2 * v / (pi * width^4 * factorial(j)) * exp(-v^2 / width^2) * (v^2 / width^2)^(j - 1) * (j - (v^2 / width^2))
end
quad_samples, quad_weights = gausslegendre(100);
# Transform from [-1, 1] to [0, 20]
v_samples = @. 10.0 * (1.0 + quad_samples)
v_weights = @. 10.0 * quad_weights
# Transform from [-1, 1] to [0, pi/2] for interior quadrature calculations
theta = 0.25 * pi * (1.0 .+ quad_samples)
theta_weights = 0.25 * pi .* quad_weights
function Q_s(alpha, beta)
return (2 * alpha / sin(pi * alpha)) * dot(theta_weights, besselj0.(2 * beta * cos.(theta)) .* cos.(2 * alpha .* theta))
end
function Qp_s(alpha, beta)
return -(4 * alpha / sin(pi * alpha)) * dot(theta_weights, besselj1.(2 * beta * cos.(theta)) .* cos.(theta) .* cos.(2 * alpha .* theta))
end
function R22(alpha, beta)
return (-beta^2 / sin(pi * alpha)) * dot(theta_weights, cos.(2 * alpha * theta) .* (besselj.(2, 2 * beta * cos.(theta)) - besselj0.(2 * beta * cos.(theta)) .* cos.(2 * theta)))
end
# quadgk integration tolerance
rtol = 1e-10
function r11(w)
alpha = -w * wpwc
function integrand(v)
beta = -knorm * v / v_0
return dF0_dv(v) * alpha * (1 - Q_s(alpha, beta))
end
I, err = quadgk(integrand, 0.0, Inf, rtol=rtol)
return I
end
function r12(w)
alpha = -w * wpwc
function integrand(v)
beta = -knorm * v / v_0
return -dF0_dv(v) * 0.5 * im * beta * Qp_s(alpha, beta)
end
I, err = quadgk(integrand, 0.0, Inf, rtol=rtol)
return I
end
function r22(w)
alpha = -w * wpwc
function integrand(v)
beta = -knorm * v / v_0
return -dF0_dv(v) * (-beta^2 / sin(pi * alpha)) * dot(theta_weights, cos.(2 * alpha * theta) .* (besselj.(2, 2 * beta * cos.(theta)) - besselj0.(2 * beta * cos.(theta)) .* cos.(2 * theta)))
end
I, err = quadgk(integrand, 0.0, Inf, rtol=rtol)
return I
end
function dispersion(w)
coeff = 2.0 * pi * v_0^2 * wpwc / (knorm^2 * w)
x11 = coeff * r11(w)
x12 = coeff * r12(w)
x22 = coeff * r22(w)
k11 = 1.0 + x11
k12 = x12
k21 = -x12
k22 = 1.0 + x22
D = k11 * (k22 + knorm^2 / (v_0 ^2 * w^2)) - k12 * k21
return D
end
w = wr + im * wi
return dispersion(w)
end
function solve_es_dispersion(;knorm, j, width, wpwc, wr, wi)
end
function plot_contours_from_file(filename)
fid = HDF5.h5open(filename, "r")
x = HDF5.read(fid["w_r"])
y = HDF5.read(fid["w_i"])
D = HDF5.read(fid["D"])
case_id = HDF5.read(fid["caseId"])
wpwc = HDF5.read(fid["wpwc"])
close(fid)
z = abs.(D)
min_log = minimum(z) / maximum(z)
min_level = trunc(Int, log(min_log))
levels = -min_level:0
colorbar_labels = [L"10^{%$i}" for i in levels]
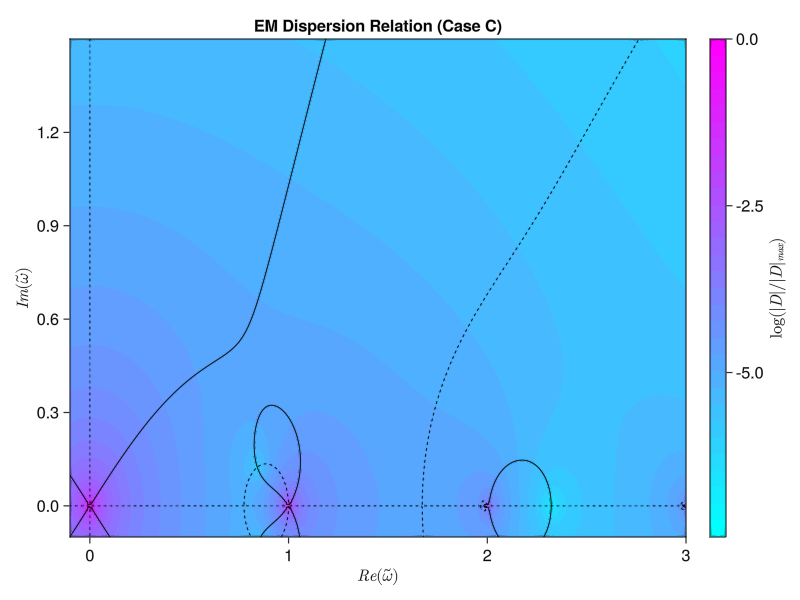
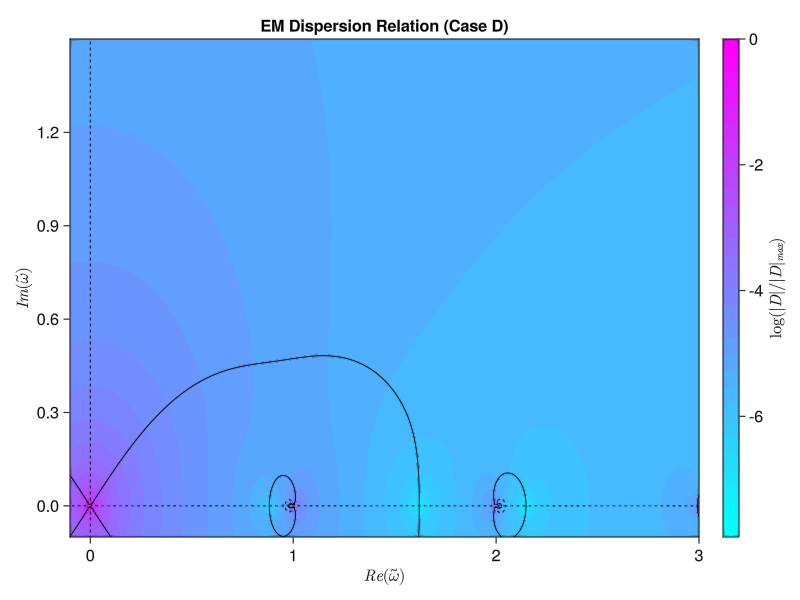
plt = contourf(x, y, log10.(z ./ maximum(z)), levels=30, color=:cool, colorbar_ticks=(levels, colorbar_labels))
title!(L"$|D|/|D|_{max}$ (Case " * case_id * ")")
xlabel!(L"Re(\tilde{\omega})")
ylabel!(L"Im(\tilde{\omega})")
savefig(plt, "julia-dgh-case" * case_id * ".pdf")
display(plt)
contour_plt = contour(x, y, real.(D), levels=[0.0, 0.001], c=[:black], ls=[:solid])
contour!(contour_plt, x, y, imag.(D), levels=[0.0, 0.001], c=[:black], ls=[:dash])
title!(L"$Re(D)=0$ and $Im(D)=0$ (Case " * case_id * ")")
display(contour_plt)
end
function run_case(case, file_base_name, num_pts)
filename = file_base_name * "-" * case.id * ".h5"
fid = HDF5.h5open(filename, "w")
fid["caseId"] = case.id
fid["knorm"] = case.knorm
fid["j"] = case.j
fid["width"] = case.width
fid["wpwc"] = case.wpwc
x = range(-0.1, 3.0, length=num_pts)
y = range(-0.1, 1.5, length=num_pts)
fid["w_r"] = collect(x)
fid["w_i"] = collect(y)
function em_dispersion(wr, wi)
return solve_em_dispersion(knorm=case.knorm, j=case.j, width=case.width, wpwc=case.wpwc, wr=wr, wi=wi)
end
println("Running case ", case.id, " over ", num_pts^2, " total points..." )
D = @. em_dispersion(x' ./ case.wpwc, y ./ case.wpwc)
fid["D"] = D
close(fid)
println("Finished. Writing to file ", filename)
return filename
end
filename = run_case(cases[1], "data", 320)
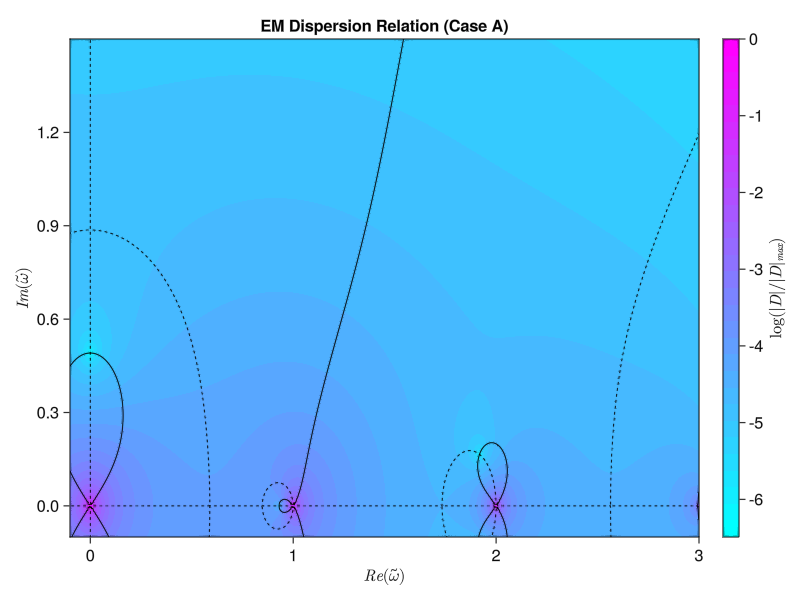
plot_contours_from_file(filename)It’s very close! The basic anatomy looks to be nearly correct. Most importantly, the fastest-growing mode (the zero with the largest imaginary frequency component) that we see appears to be near the expected $\tilde{\omega} = 0.0 + 0:4912i$:
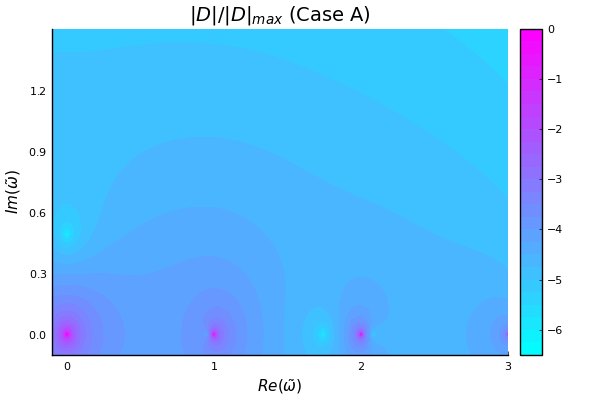
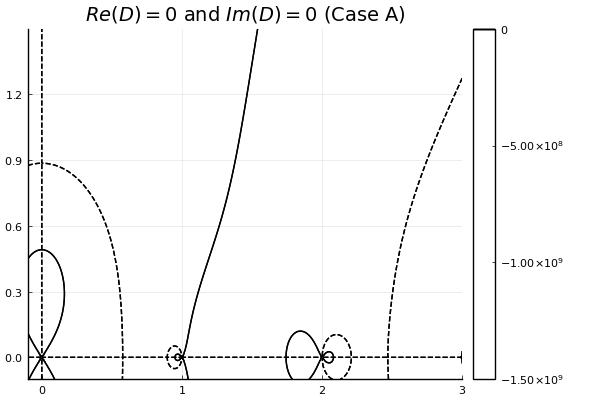
We can confirm by running a minimization scheme near the zero to get a precise value. I’ll use the Optim.jl library to perform the descent minimization:
function find_zero(case)
function em_dispersion(w)
return abs(solve_em_dispersion(knorm=case.knorm, j=case.j, width=case.width, wpwc=case.wpwc, wr=w[1], wi=w[2]))
end
println("========== Case ", case.id, " ==========")
for guess in case.guesses
start = [guess[1] / case.wpwc, guess[2] / case.wpwc]
println("Searching for zero near w = ", guess[1], " + ", guess[2], "i")
result = optimize(em_dispersion, start, Optim.Options(x_tol = 1e-6))
println("Case ", case.id, ":")
display(result)
println("Did we find a root? Maybe! We think there is a root at:")
println("w = ", round(Optim.minimizer(result)[1] * case.wpwc, digits=5), " + ", round(Optim.minimizer(result)[2] * case.wpwc, digits=5), "i")
end
end
find_zero(cases[1])========== Case A ==========
Searching for zero near w = 0.0 + 0.5i
* Status: success
* Candidate solution
Final objective value: 2.739900e-08
* Found with
Algorithm: Nelder-Mead
* Convergence measures
√(Σ(yᵢ-ȳ)²)/n ≤ 1.0e-08
* Work counters
Seconds run: 3 (vs limit Inf)
Iterations: 94
f(x) calls: 184
Case A:
Did we find a root? Maybe! We think there is a root at:
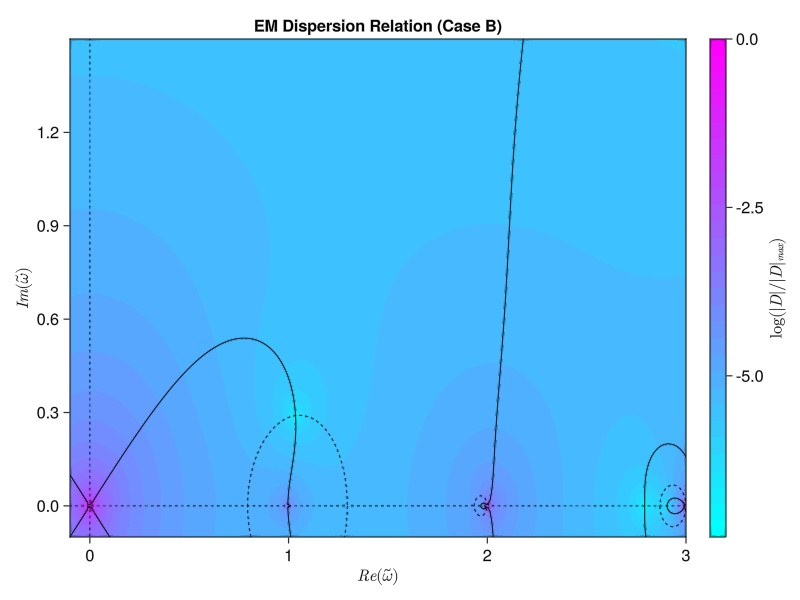
w = 0.0 + 0.49123iSo far so good! But I am very suspicious that I don’t see any other growing modes. From Iman’s paper I would expect to see another solution near $\tilde{\omega} = 1.9 + 0.2i$ Let’s try a different case, setting $\tilde{k} = 4.65$. This is “Case B” in the paper, and I expect a solution at $\tilde{\omega} = 1.0363 + 0.2900i$:
========== Case B ==========
Searching for zero near w = 1.0 + 0.3i
* Status: success
* Candidate solution
Final objective value: 2.216717e-08
* Found with
Algorithm: Nelder-Mead
* Convergence measures
√(Σ(yᵢ-ȳ)²)/n ≤ 1.0e-08
* Work counters
Seconds run: 3 (vs limit Inf)
Iterations: 89
f(x) calls: 175
Case B:
Did we find a root? Maybe! We think there is a root at:
w = 1.03461 + 0.28968iHmm, that is much further off than I would like. Decreasing the tolerance for the QuadGK integrals does not change the result, and increasing the number of Gauss-Legendre quadrature points also makes no difference.
How about we try doing the v integrals by quadrature as well?
No quadrature, two contour plots with 400pts each: 16.7 sec Quadrature, two contour plots with 400pts each: 6.1 sec!
Back to the original attempt code, evaluating with 1600pts each and threading I get 30.7s with QuadGK, and 6.3s with my own quadrature! Let’s definitely use that instead:
function solve_em_dispersion(; knorm, j, width, wpwc, w_array)
# All normalization parameters
Ze = -1.0 # Charge ratio for species
Ae = 1.0 # Mass ratio for species
wptau = 1.0 # Time normalization
wctau = 1.0 / wpwc # omega_c * tau
Omega = Ze / Ae / wpwc # Species-dependent cyclotron frequency
B0z = 1.0 # Magnetic field magnitude
vp0 = sqrt(2.0) # Ring distribution peak velocity
k = knorm / vp0 / wpwc # Normalized k
ne = 1.0 # Species density
wce = Ze / Ae * B0z # Normalized cyclotron frequency
wp2 = Ze^2 * ne / Ae # Normalized plasma frequency squared
beta = @. k * v_perp / wce / wctau
f0prime = @. 2 * v_perp / (pi * width^4 * factorial(j)) * exp(-v_perp^2 / width^2) * (v_perp^2 / width^2)^(j - 1) * (j - (v_perp^2 / width^2))
Q = similar(beta, ComplexF64) # Initialize empty Q
Qprime = similar(beta, ComplexF64) # Initialize empty Q'
jprime_ma_jprime_pa = similar(beta, ComplexF64) # Initialize empty J'_-a(b)*J'_+a(b)
D = similar(w_array, ComplexF64) # Output array for each input w
for (idx, w) in enumerate(w_array)
alpha = w / wce / wctau
sine_factor = pi * alpha / sin(pi * alpha)
cos_2_a_theta = @. cos(2 * alpha * theta)
for ii in 1:length(beta)
two_beta_cos_theta = (2.0 * beta[ii]) .* cos_theta
Q[ii] = sine_factor * 2.0 / pi * sum(theta_weights .* (besselj0.(two_beta_cos_theta) .* cos_2_a_theta))
Qprime[ii] = sine_factor * -4.0 / pi * sum(theta_weights .* (besselj1.(two_beta_cos_theta) .* cos_theta .* cos_2_a_theta))
jprime_ma_jprime_pa[ii] = 1.0 / pi * (sum(theta_weights .* (cos_2_a_theta .* (besselj.(2, two_beta_cos_theta) .- besselj0.(two_beta_cos_theta) .* cos_2_theta))) - alpha .* sin.(pi .* alpha)/ beta[ii]^2)
end
v_integrand_11 = @. - f0prime * (alpha / beta^2 * (1.0 - Q)) * v_perp^2
v_integrand_12 = @. f0prime * (im / 2.0 / beta * Qprime) * v_perp^2
v_integrand_22 = @. f0prime * (sine_factor / alpha * jprime_ma_jprime_pa + alpha / beta^2) * v_perp^2
coeff = wptau^2 / wctau * 2.0 * pi * wp2 / w / wce
chi_xx = coeff * sum(v_weights .* v_integrand_11)
chi_xy = coeff * sum(v_weights .* v_integrand_12)
chi_yy = coeff * sum(v_weights .* v_integrand_22)
K_xx = 1.0 + chi_xx
K_xy = chi_xy
K_yx = - K_xy
K_yy = 1.0 + chi_yy
D[idx] = K_xx * (K_yy - wptau^2 / w^2 / wctau^2 * k^2) - K_xy * K_yx
end
return D
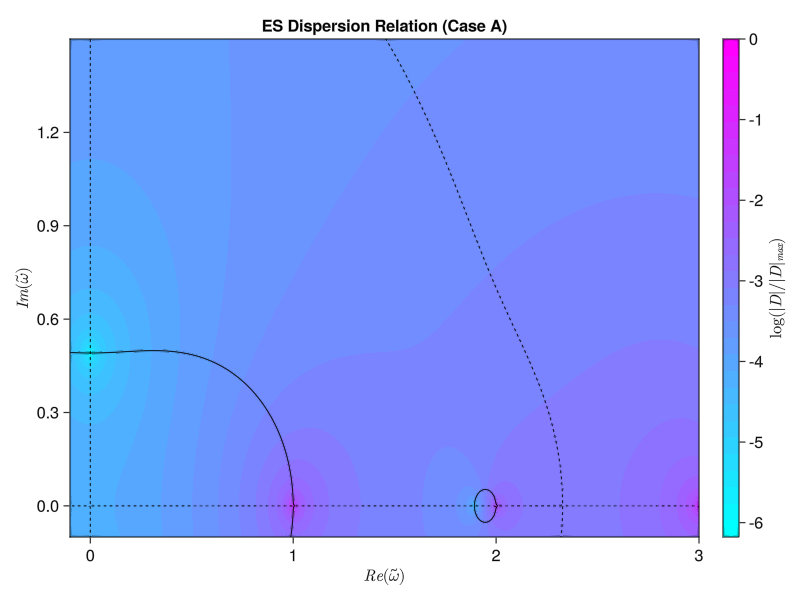
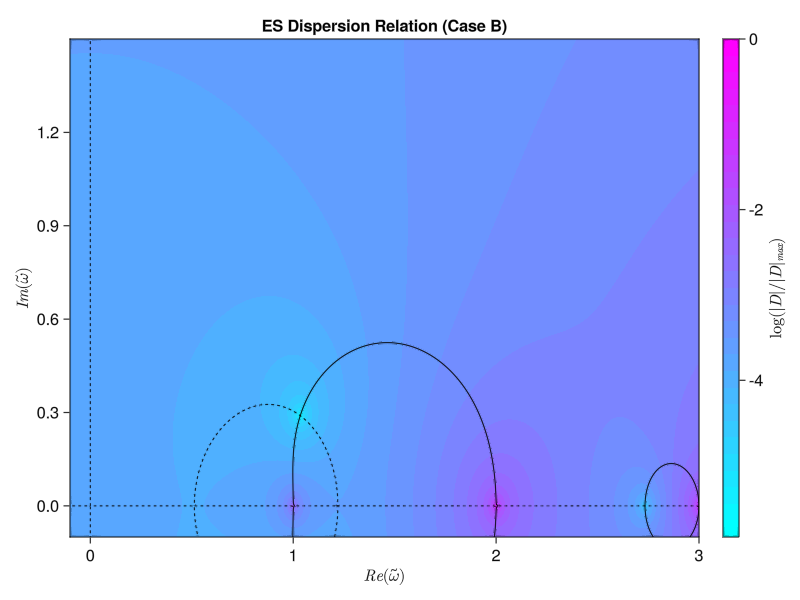
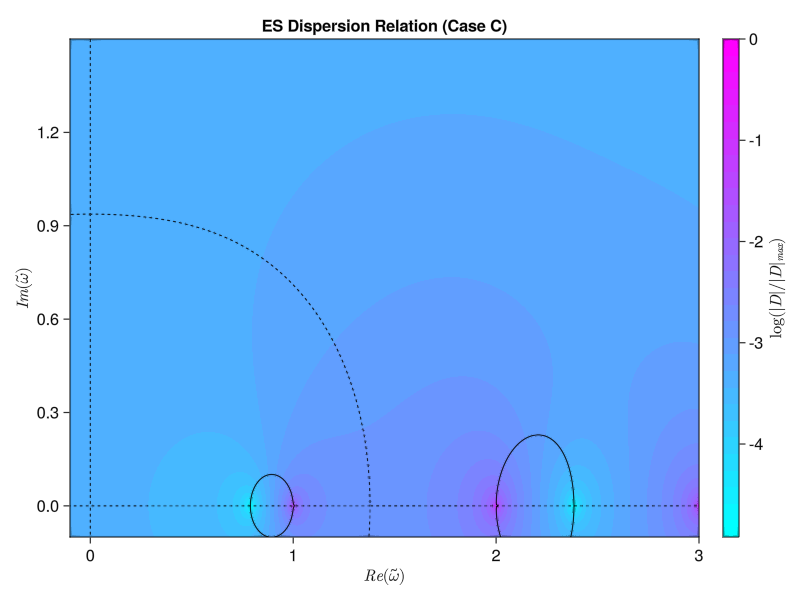
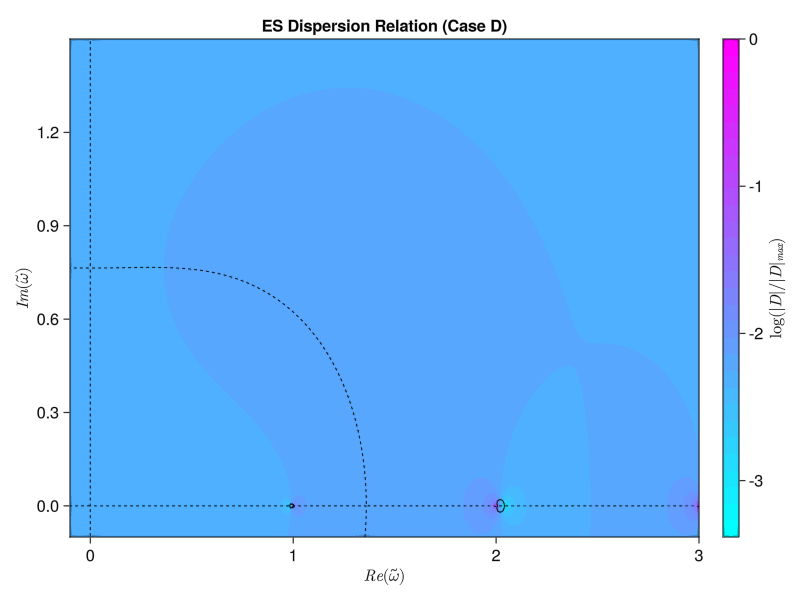
endWe’ve got to use some for loops instead of straight broadcasting to do the double integral properly, but now we get the same results as the paper! Similarly, we should be able to compute the electrostatic version using either the simplified single-species version from Tartaronis or the full multi-species version from Vogman. We’ll use the latter:
\[D(\omega, k_\perp) = 1 + \sum_s M_s \frac{\omega_p ^2}{\omega_c ^2} \int_0 ^\pi \frac{\sin(\omega \tau / \Omega_{cs})}{\sin(\omega \pi / \Omega_{cs})} \sin (\tau) F_0 (\tau + \pi) \dd \tau = 0\] where \[F_0 (\tau) = \int_0 ^\infty f_{s, 0} (v_\perp) J_0 \left( 2 \frac{k_\perp v_\perp}{\Omega_{cs}} \sin(\frac{\tau}{2})\right) 2 \pi v_\perp \dd v_\perp\] \[\Omega_{cs} \equiv \frac{Z_s \omega_c}{M_s \omega_p}\]
function solve_es_dispersion(; knorm, j, width, wpwc, w_array)
# All normalization parameters
Ze = -1.0 # Charge ratio for species
Ae = 1.0 # Mass ratio for species
wptau = 1.0 # Time normalization
wctau = 1.0 / wpwc # omega_c * tau
Omega = Ze / Ae / wpwc # Species-dependent cyclotron frequency
B0z = 1.0 # Magnetic field magnitude
vp0 = sqrt(2.0) # Ring distribution peak velocity
k = knorm / vp0 / wpwc # Normalized k
ne = 1.0 # Species density
wce = Ze / Ae * B0z # Normalized cyclotron frequency
wp2 = Ze^2 * ne / Ae # Normalized plasma frequency squared
# Initial distribution
vp_alpha = v_perp ./ width
f0 = @. ne / (pi * width^2 * factorial(j)) * (vp_alpha)^(2*j) * exp(- vp_alpha ^2)
z = @. k * v_perp / Omega / B0z
D = similar(w_array, ComplexF64)
for (idx, w) in enumerate(w_array)
w_alpha = w / Omega / B0z
tau_int = similar(z, ComplexF64)
for ii in 1:length(z)
# tau = range(0.0, pi, 100)
tau_integrand = @. sin(w_alpha * tau) / sin(w_alpha * pi) * sin(tau) * besselj0(2 * z[ii] * sin((tau + pi)/2))
# tau_int[ii] = trapz(tau, tau_integrand)
tau_int[ii] = sum(tau_weights .* tau_integrand)
end
v_integrand = @. f0 * tau_int * v_perp
D[idx] = 1.0 + 2.0 * pi * Ze^2 / Ae / Omega^2 * sum(v_weights .* v_integrand)
end
return D
end| Electromagnetic | Electrostatic |
|---|---|
|
|
|
|
|
|
|
|